Summary
- What Is It? – Simulated Annealing program for studying crystal defect formation on 2D manifolds
- Technologies Used – Julia
-
My Accomplishments
- Improved algorithm speed by 40x
- Drove project to completion
- Effectively communicated technical requirements with a non-technical stakeholder
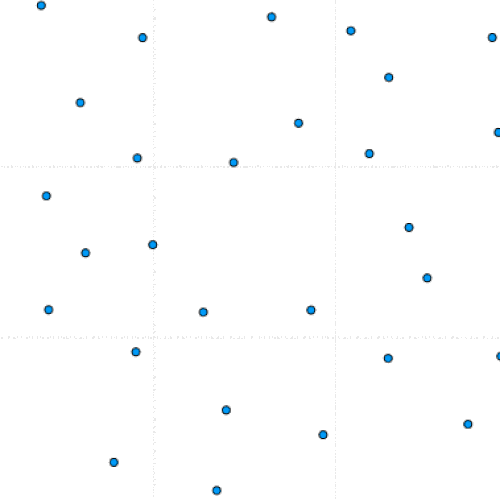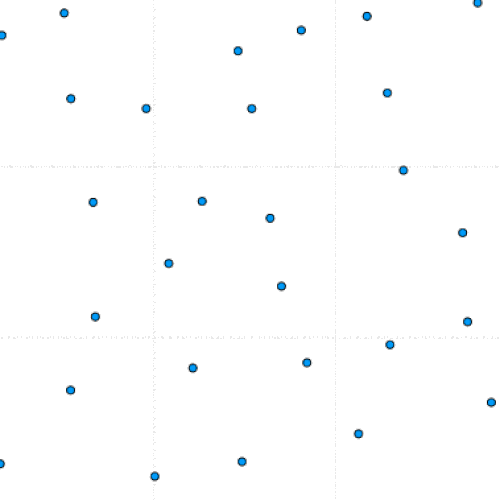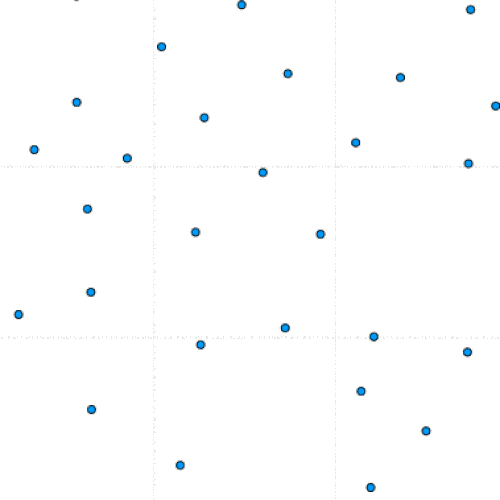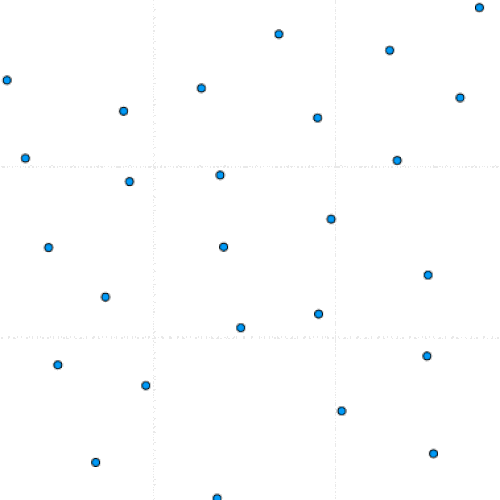
This project is part of a team effort to study colloids movement on 2D manifolds, the formation of lattice defects, and how they relate to the morphing of viral capsids. The team was led by Dr. Luigi Perotti at the University of Central Florida.
What it Does
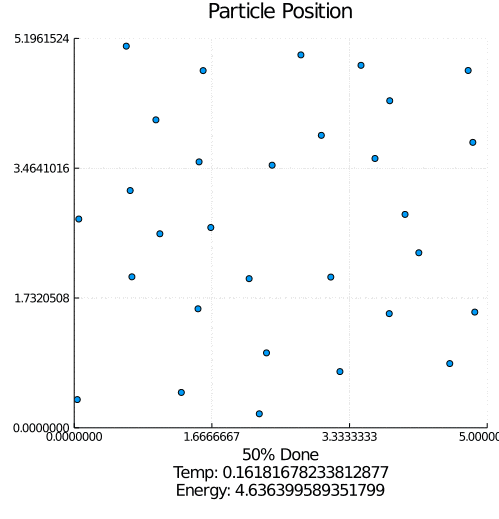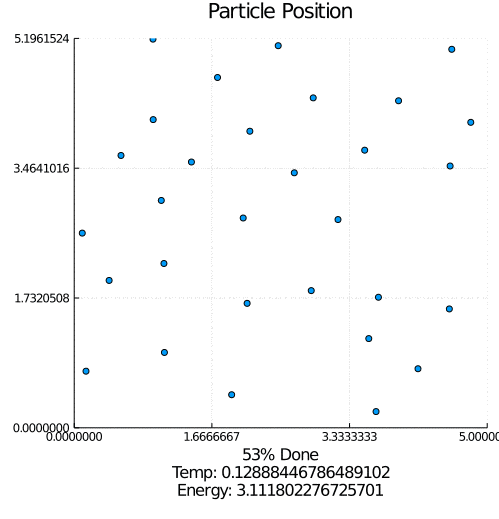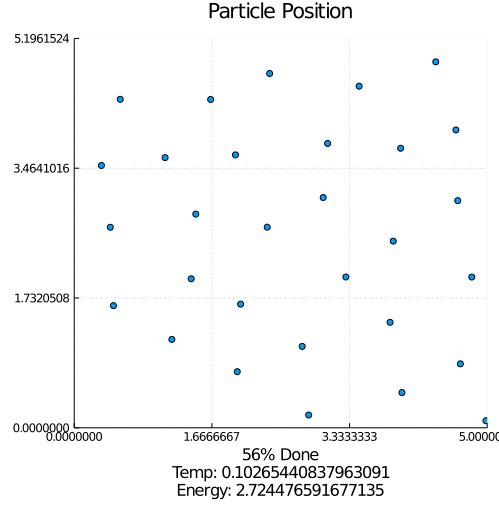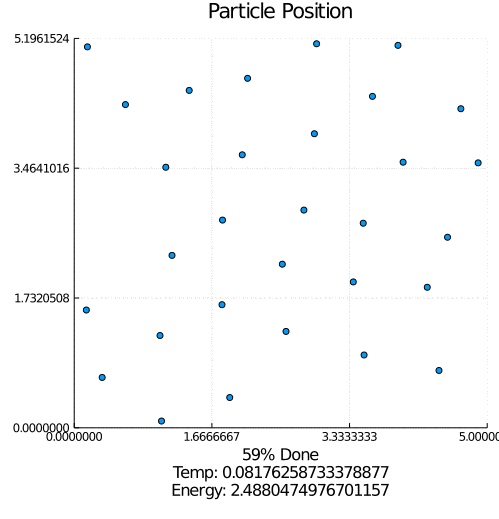
This program simulates particle interaction using a modified version of the Kinetic Monte Carlo Algorithm. After taking in a number of parameters including the number of particles, the number of iterations, and the equilibrium distance, the program initializes a set of randomly positioned particles.
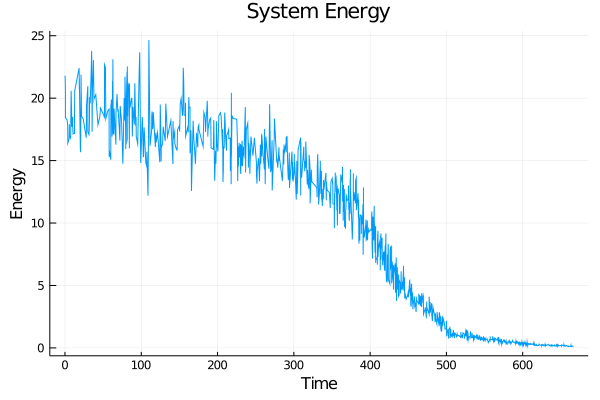
During each timestep, each particle is perturbed and this move is either saved or reverted based on whether it lowers the total system energy. Eventually, the lowest possible energy configuration is reached.
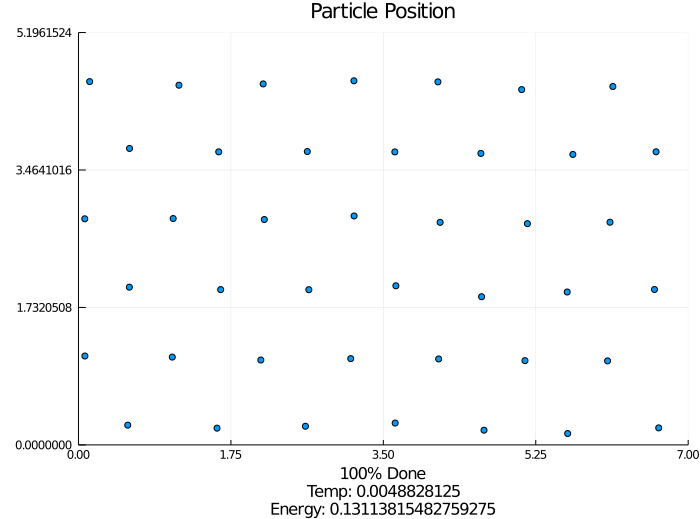
How I Did It
I started with the pseudo-code for the modified Kinetic Monte Carlo algorithm. Using the Julia programing language, I implemented the algorithm along with supporting features such as estimated completion time, outputting an energy-time graph, and creating a particle movement animation. This iteration was extremely slow, but it worked.
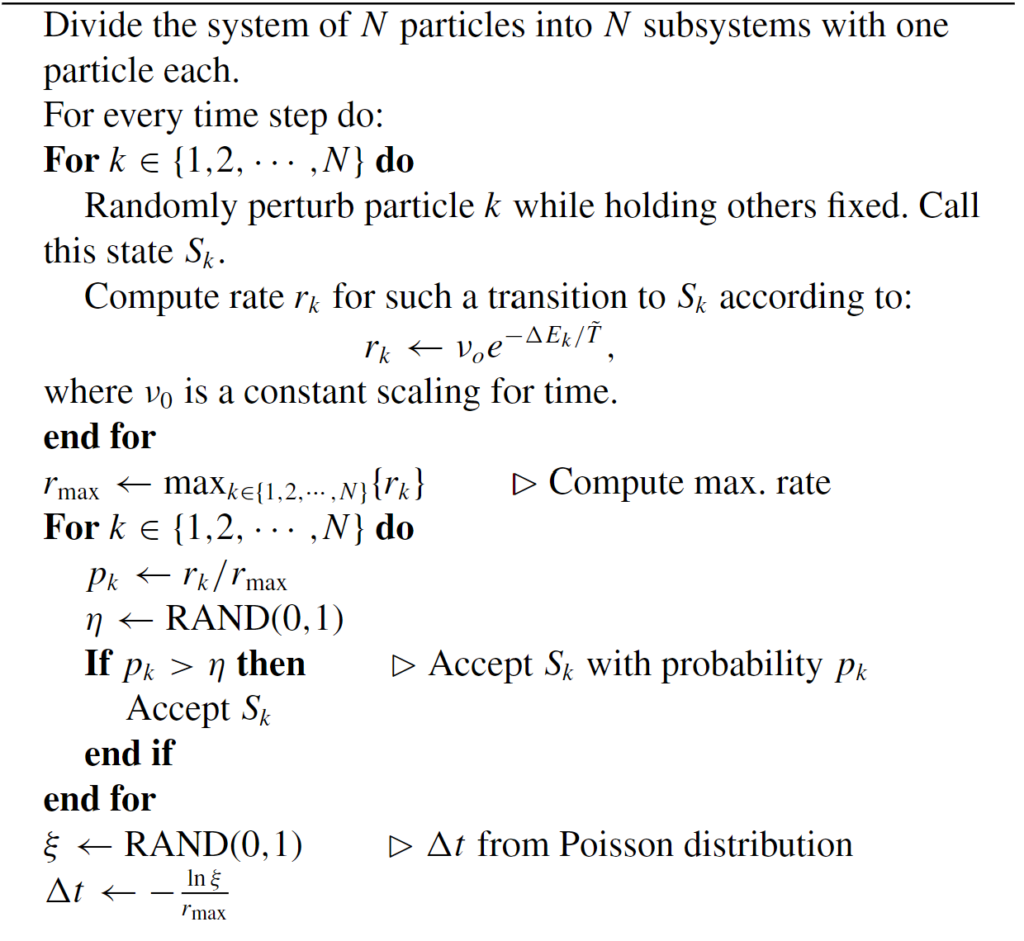
Improvements I Made
I was able to double the speed of my program simply by reducing repeated computations. For example, if I calculate the distance between particle a and particle b, then I don’t need to compare the distance again between b and a. I was also able to utilize parallelization, as each particle movement is independent.
From here I completely restructured the entire program to improve the energy calculation time complexity from O(n2) to O(n). This was a huge improvement because it increase the maximum number of particles from around 30 to a couple thousand. I did this by spatially encoding the position of each particles in memory. I broke the region up into zones and each zone was an independent particle array. Each particle only needs to check it’s own zone, and the eight adjacent zones. Because zones scale with equilibrium distance, each particle only needs to compare to a small constant number of other particles.
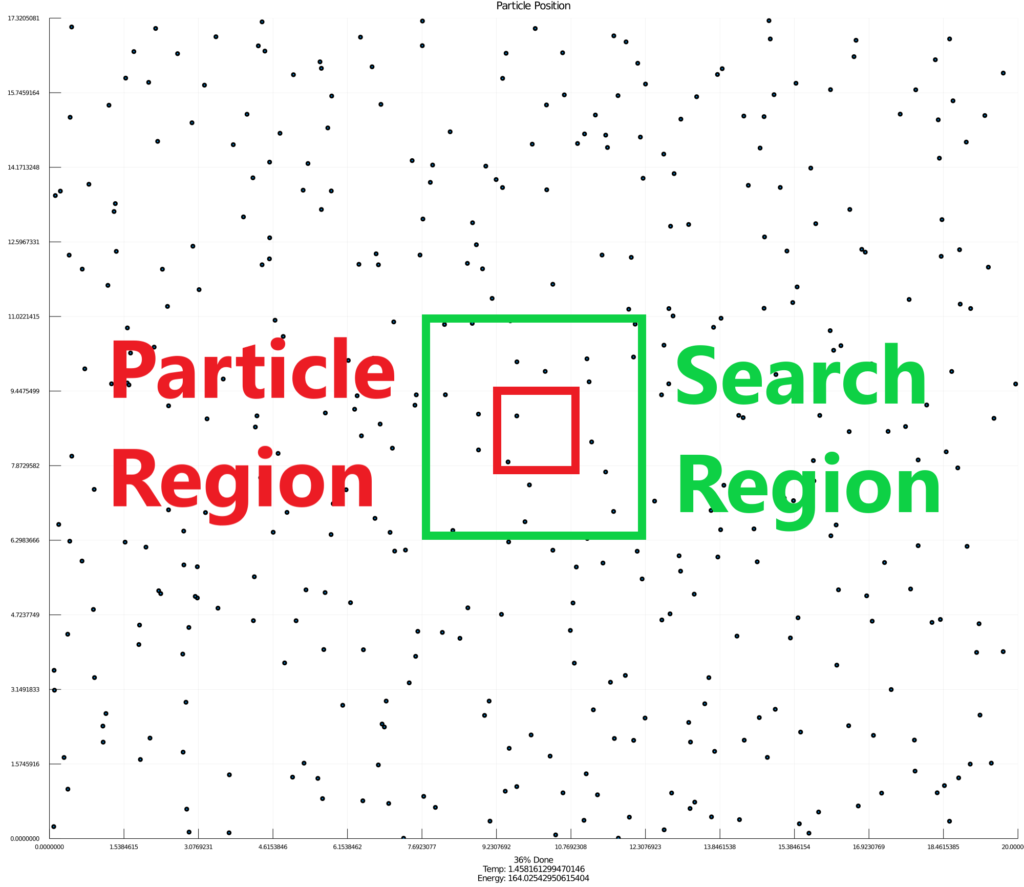
Overall, the improvements I made were able to speed up the original program by over 40 times.
What I Learned
This was the most challenging project I have ever completed and I learned so much during the process. First, I had never worked with this programming language before, but now I would consider myself proficient in Julia. Another thing I learned was how to obtain technical requirements from a non-technical person. I was given the task to “implement this algorithm” and had to figure everything out on my own. This taught me how to ask questions and learn what the goal of the project was, as well as determine what features would contribute to the goal. In implementing parallelism and my spatial encoding scheme, I learned a lot about memory management and working with shared memory. The final big lesson was learning how to develop software as a piece of a bigger project. My program need to accept inputs from a completely separate program and then map outputs in a way the next program would understand. Communicating with my team and being able to work together to line everything up was definitely challenging, but I developed a deeper understanding of team dynamics in the process.